Four half-day tutorials will be presented on Tuesday April 18, 2017. Details of each tutorial are provided below.
Chairs
- Andrew Zalesky, The University of Melbourne, Australia
- Oscar Acosta, University Rennes 1, France
 Tutorial 1: Neuroimaging analysis within R
Tutorial 1: Neuroimaging analysis within R
Organizers
Dr John Muschelli, Johns Hopkins Bloomberg School of Public Health, USA
Dr Kristin Linn, University of Pennsylvania, USA
Overview
In this tutorial, we will provide tutorials on how to use R for structural magnetic resonance imaging (MRI) analysis. We will show how to perform entire image analysis in R, from the scans in raw image format to the statistical analysis after image preprocessing, with an emphasis on reproducibility by using a single programming language. In this hands-on tutorial, attendees will be given instructions for setup and data before the course, so that they are able to follow along and perform the analysis during the tutorial.
Topics
- Introduction to the Statistical Software R (JM)
- Read and Write Images (JM)
- Visualization (JM and JPF)
- Inhomogeneity Correction (JPF)
- Brain Extraction (JM)
- Image Segmentation (JPF)
- Coregistration Within and Between MRI Studies (JM)
- Intensity Normalization (JPF)
- Harmonization of mutli-site datasets (JPF)
Audience
The intended audience for this tutorial is broadly researchers and analysts working on structural MRI analysis. Prior knowledge of programming in R is useful, but not absolutely necessary.
 Tutorial 2: Biomedical texture analysis
Tutorial 2: Biomedical texture analysis
Organizers
Dr Adrien Depeursinge, EPFL / HES-SO Valais, Switzerland
Dr Yang Song, University of Sydney, Australia
Dr Weidong (Tom) Cai, University of Sydney, Australia
Co-presenters
Dr Fan Zhang, Brigham & Women’s Hospital, Harvard Medical School, USA
Dr Wojciech Chrzanowski, University of Sydney, Australia
Dr Yong Xia, Northwestern Polytechnical University, China
Overview
This tutorial will provide an overview of recent developments in the field of biomedical texture analysis. Texture based imaging biomarkers complement focal, invasive biopsy based biomarkers by providing information on tissue structure over broad regions, non-invasively. Texture has been used to predict patient survival, tissue function, disease subtypes and genomics. Nevertheless, several challenges remain, such as: the lack of an appropriate framework for multi-scale, multi-spectral analysis in 2D and 3D; localization of uncertainty of texture operators; validation; and, translation to routine clinical applications.
This tutorial will establish the mathematical foundations of multidimensional texture operators and will review the limitations of popular methods, including deep convolutional neural networks. We will describe recent research to address the major challenges outlined above. High-impact potential clinical applications will be presented by several experts in the fields of radiology, neuroimaging and pharmaceutical research.
Topics
Introduction (Adrien Depeursinge, Yang Song, Weidong Cai)
- Image processing
1 Fundamentals and review of digital texture processing methods for biomedical image analysis (Adrien Depeursinge)
1.2 Texture analysis methods for different biomedical imaging modalities (Yang Song) - Applications (Invited speakers)
1 Diagnosis of brain disorders based on textures in neuroimaging (Fan Zhang)
2.2 Texture analysis for image-based computer-aided diagnosis (Yong Xia)
2.3 Image texture information in pharmaceutical research (Wojciech Chrzanowski)
Closing remarks (Adrien Depeursinge, Yang Song, Weidong Cai)
Audience
The target audience is general ISBI attendees with basic knowledge in image processing. Researchers who are interested in biomedical texture analysis of various imaging modalities (e.g. CXR, CT, MRI, PET-CT, microscopy) and applications (e.g. disease classification, detection) are welcome to join the tutorial.
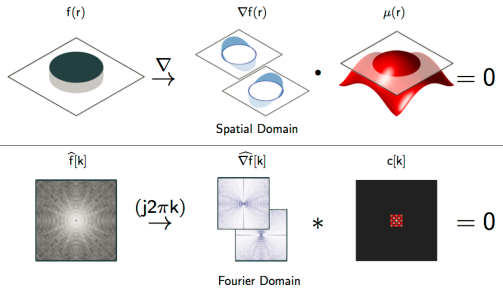 Tutorial 3: Continuous domain sparse recovery of biomedical image data using structured low-rank approaches
Tutorial 3: Continuous domain sparse recovery of biomedical image data using structured low-rank approaches
Organizers
Professor Jong Chul Ye, KAIST, South Korea
Professor Mathews Jacob, University of Iowa
Overview
In this tutorial, we will provide a detailed review of recent advances in structured low-rank matrix completion formulation, where the recovery of a continuous domain image from its sparse measurements is considered. This framework is centered on the fundamental duality between the sparsity of the continuous signal in the spatial domain and the low-rankness of a structured matrix in the spectral (Fourier) domain. This property enables us to reformulate sparse recovery of continuous domain signal as a low-rank matrix completion problem in the spectral domain, thus providing the benefit of sparse recovery with performance guarantees. The proposed low-rank completion approach can be regarded as a generalization of recent spectral compressed sensing to recover large classes of finite rate of innovations (FRI) signals at near optimal sampling rates. We will also review fast algorithms that are comparable in complexity to current compressed sensing methods, which enable the application of the framework to large scale biomedical image recovery problems. We will demonstrate the utility of the framework in a wide range of biomedical imaging applications such as compressed sensing MRI, super-resolution microscopy, ultrasound, as well as other image processing applications.
Topics
- Introduction
- Review of compressed sensing
- FRI signal recovery from uniform samples
3.1. Review of 1-D FRI sampling theory
3.2. 2-D FRI: Generalization to multidimensional signals - Structured low-rank interpolation for non-uniform samples
4.1. 1-D Theory
4.2. 2-D Theory - Fast Implementations
- Biomedical imaging applications
6.1. MRI – Part 1
6.1.1. Super-resolution MRI
6.1.2. CS-MRI using structured low-rank penalty
6.2. MRI – Part 2
6.2.1. CS MRI as a low-rank interpolation
6.2.2. MR artifact removal
6.3. Other biomedical imaging applications: super-resolution microscopy, scanning microscopy, X-ray CT, ultrasound imaging
Audience
The target audience is general ISBI attendees with basic knowledge and interest in image recovery or restoration. Researchers who are interested in the reconstruction/restoration of MRI, CT, Ultrasound, Microscopy are welcome to join the tutorial.
 Tutorial 4A: Deep learning based feature detection using DIGITS
Tutorial 4A: Deep learning based feature detection using DIGITS
Organizers
Mike Wang, NVIDIA
Dr Jason Rigby, Monash University
Dr Wojtek Goscinski, Monash University
Overview
Extracting salient features for detailed analysis from medical images (such as X-ray, MRI, CT, etc) can be a laborious and time-consuming task. Where good quality tagged training data exists, Deep Neural Networks (DNNs) have been very successfully applied to accomplish this task in recent years. This tutorial explore introduces several approaches to image classification and feature detection within images using DNNs.
You will learn through a hands-on exercise how to use the NVIDIA DIGITS web interface-based system to perform both training and inference tasks. We will use an open retinal imaging dataset during the tutorial. The web interface includes a Python interpreter, which we will use to perform some basic image preparation prior to the training.
To take part in the hands-on exercise, you need a laptop with a working internet connection with a websockets-capable browser installed (recent version of Chrome or Firefox recommended). Please test your browser using the following link: https://websocketstest.com/. No further software is required.
For further information about DIGITS: Deep Learning GPU Training System please visit: https://devblogs.nvidia.com/parallelforall/digits-deep-learning-gpu-training-system/
![]() Tutorial 4B: The Frontier concept and integration within syngo.via
Tutorial 4B: The Frontier concept and integration within syngo.via
Presenters
Dr Anders Rodell, Siemens Healthineers ANZ
Overview
An ideal research environment gives you access to the latest applications, provides tools that translate your ideas into tangible prototypes, and supports your exchange with other experts around the world.
With syngo.via Frontier, you can explore the potential of advanced post-processing prototypes that are seamlessly integrated with your routine syngo.via system. A syngo.via Frontier Development Kit option also enables you to easily implement your own algorithms and connects you directly with other key opinion leaders and the Siemens Healthcare pre-development teams. Save time and reduce costs with an integrated research solution. Boost your reputation and attract talent as well as patients. Bridge the gap in post-processing translational research with syngo.via Frontier.
Define individual tools for your clinical research and overcome obstacles like stand-alone programs that are frequently difficult to integrate in a hospital environment. The syngo.via Frontier Development Kit provides you with an integrated solution for seamlessly translating individualised advanced post processing requirements into tangible prototypes which can be deployed the clinical infrastructure.
Get started with the prototype Starter Kit that enables you to easily reuse existing code for your own prototype development.
Topics
- Theoretical introduction to:
1.1 The Frontier concept and integration within syngo.via
1.2 The Frontier processing prototypes currently available
1.3 The Frontier Development and Starter Kit - Practical demonstration of:
2.1 Data handling within Frontier
2.2 Select Frontier prototypes
2.3 The Mevislab Development environment for Frontier

